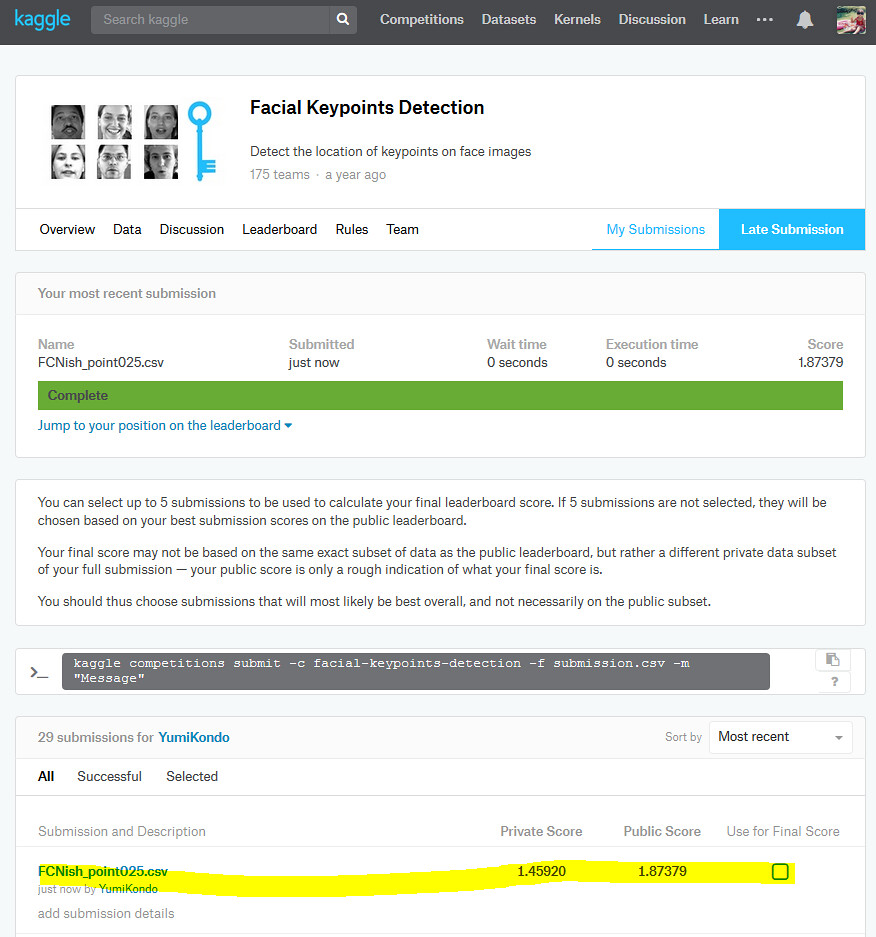
My Kaggle's final score using the model explained in this ipython notebook.
from IPython.display import IFrame
src = "https://www.youtube.com/embed/8FdSHl4oNIM"
IFrame(src, width=990/2, height=800/2)
A few days ago, I saw this very awesome youtube video demonstrating the performance of the state-of-art facial keypoint detection. This video was a part of ICCV 2017's presentation How far are we from solving the 2D & 3D Face Alignment problem? (and a dataset of 230,000 3D facial landmarks).
This paper says that the current state-of-art landmark localization problem uses Hour-Glass network of Stacked Hourglass Networks for Human Pose Estimation. Hour-Glass network uses the idea from the fully convolutional networks, which is often used in object segmentation task. I also studied FCN in previous blog using object segmentation data. Here, I was originally confused about how to apply the model from the object segmentation task to the facial landmark detection task. The data structure of the two problems are quite different:
- facial landmark detection:
- input: image
- output: x,y coordinate of landmarks
- object segmentation:
- input: image
- output: the object's class at every pixcel
For example in my previous blog, Achieving Top 23% in Kaggle's Facial Keypoints Detection with Keras + Tensorflow, I did a facial keypoints (landmarks) detection using Kaggle's facial keypoints detection data. In this post, I used CNN to extract features and then regress on the x,y coordinates of the landmarks.
So how can we apply the model from the object segmentation to the facial landmark detection problem? The answer was in the preprocessing of images: the (x,y)-coordinates of the landmarks are transformed to "heatmap" using some kernels e.g. Gaussian kernel. Then the problem becomes estimating the value of the heatmap at every pixcel just like object detection problem where the goal is to estimate the object's class at every pixcel. Interesting!
In this blog post, I will revisit Kaggle's facial keypoints detection data to learn the performance of simple FCN-like model for the facial keypoint detection problem.
I was able to improve the private and public scores of this competition from my previous model. The script here can yield:
- Private score : 1.45920
- Public score : 1.87379
This means that I am "roughly" in Top 5 in the private score and Top 6 in the public score out of 175 teams. I use the word "roughly" because the competition has ended in January 2017, and final scores are not available for me. (The final offical scores are evaluated using 50% of the testing data while the results here are based on ALL the testing data. Nevertheless, the model performance is pretty good.
Import libraries¶
import matplotlib.pyplot as plt
import tensorflow as tf
from keras.backend.tensorflow_backend import set_session
import keras, sys, time, os, warnings, cv2
from keras.models import *
from keras.layers import *
import numpy as np
import pandas as pd
warnings.filterwarnings("ignore")
os.environ["CUDA_DEVICE_ORDER"] = "PCI_BUS_ID"
config = tf.ConfigProto()
config.gpu_options.per_process_gpu_memory_fraction = 0.95
config.gpu_options.visible_device_list = "1"
set_session(tf.Session(config=config))
print("python {}".format(sys.version))
print("keras version {}".format(keras.__version__)); del keras
print("tensorflow version {}".format(tf.__version__))
The data is downloaded from Kaggle's facial keypoints detection, and saved under data folder.
FTRAIN = "data/training.csv"
FTEST = "data/test.csv"
FIdLookup = 'data/IdLookupTable.csv'
Data importing and preprocessing¶
Create a Gaussian heatmap¶
I will use the data loading functions from Achieving Top 23% in Kaggle's Facial Keypoints Detection with Keras + Tensorflow with some modifications that transform each (x,y)-coordinate facial keypoint to a single heatmap using Gaussian kernel. For example, in Kaggle's facial keypoints detection problem, there are 15 facial keypoints to estimate. So this means that I will create 15 heatmaps.
In the case of left eye landmark, the Gaussian kernel centered around left eye landmark $( x_{\textrm{left eye landmark}}, y_{\textrm{left eye landmark}})$ has a value at ($x,y$) as:
$$ \frac{1}{2\pi \sigma^2 } exp \left({-\frac{(x-x_{\textrm{left eye landmark}})^2 + (y-y_{\textrm{left eye landmark}})^2}{2 \sigma^2}}\right) = constant * \left({-\frac{(x-x_{\textrm{left eye landmark}})^2 + (y-y_{\textrm{left eye landmark}})^2}{2 \sigma^2}}\right) $$
I need to pre-specify $\sigma^2$ as a hyper parameter. I found that the choice of $\sigma^2$ is important to get sensible results. If $\sigma^2$ is too low, the heatmap becomes too sparse (mostly zero) for a model to train. If $\sigma^2$ is too high, the trained model focuses too much on estimating the magnitude of non-landmark coordinates.
The constant scale is another hyper parameter that also needs to be adjusted via, e.g., cross-validation.
The following functions are for transforming (x,y)-coordinate of a landmark to a heatmap.
def gaussian_k(x0,y0,sigma, width, height):
""" Make a square gaussian kernel centered at (x0, y0) with sigma as SD.
"""
x = np.arange(0, width, 1, float) ## (width,)
y = np.arange(0, height, 1, float)[:, np.newaxis] ## (height,1)
return np.exp(-((x-x0)**2 + (y-y0)**2) / (2*sigma**2))
def generate_hm(height, width ,landmarks,s=3):
""" Generate a full Heap Map for every landmarks in an array
Args:
height : The height of Heat Map (the height of target output)
width : The width of Heat Map (the width of target output)
joints : [(x1,y1),(x2,y2)...] containing landmarks
maxlenght : Lenght of the Bounding Box
"""
Nlandmarks = len(landmarks)
hm = np.zeros((height, width, Nlandmarks), dtype = np.float32)
for i in range(Nlandmarks):
if not np.array_equal(landmarks[i], [-1,-1]):
hm[:,:,i] = gaussian_k(landmarks[i][0],
landmarks[i][1],
s,height, width)
else:
hm[:,:,i] = np.zeros((height,width))
return hm
def get_y_as_heatmap(df,height,width, sigma):
columns_lmxy = df.columns[:-1] ## the last column contains Image
columns_lm = []
for c in columns_lmxy:
c = c[:-2]
if c not in columns_lm:
columns_lm.extend([c])
y_train = []
for i in range(df.shape[0]):
landmarks = []
for colnm in columns_lm:
x = df[colnm + "_x"].iloc[i]
y = df[colnm + "_y"].iloc[i]
if np.isnan(x) or np.isnan(y):
x, y = -1, -1
landmarks.append([x,y])
y_train.append(generate_hm(height, width, landmarks, sigma))
y_train = np.array(y_train)
return(y_train,df[columns_lmxy],columns_lmxy)
Functions to extract, transfer and load data¶
These functions are very similar to the ones from Achieving Top 23% in Kaggle's Facial Keypoints Detection with Keras + Tensorflow.
def load(test=False, width=96,height=96,sigma=5):
"""
load test/train data
cols : a list containing landmark label names.
If this is specified, only the subset of the landmark labels are
extracted. for example, cols could be:
[left_eye_center_x, left_eye_center_y]
return:
X: 2-d numpy array (Nsample, Ncol*Nrow)
y: 2-d numpy array (Nsample, Nlandmarks*2)
In total there are 15 landmarks.
As x and y coordinates are recorded, u.shape = (Nsample,30)
y0: panda dataframe containins the landmarks
"""
from sklearn.utils import shuffle
fname = FTEST if test else FTRAIN
df = pd.read_csv(os.path.expanduser(fname))
df['Image'] = df['Image'].apply(lambda im: np.fromstring(im, sep=' '))
myprint = df.count()
myprint = myprint.reset_index()
print(myprint)
## row with at least one NA columns are removed!
## df = df.dropna()
df = df.fillna(-1)
X = np.vstack(df['Image'].values) / 255. # changes valeus between 0 and 1
X = X.astype(np.float32)
if not test: # labels only exists for the training data
y, y0, nm_landmark = get_y_as_heatmap(df,height,width, sigma)
X, y, y0 = shuffle(X, y, y0, random_state=42) # shuffle data
y = y.astype(np.float32)
else:
y, y0, nm_landmark = None, None, None
return X, y, y0, nm_landmark
def load2d(test=False,width=96,height=96,sigma=5):
re = load(test,width,height,sigma)
X = re[0].reshape(-1,width,height,1)
y, y0, nm_landmarks = re[1:]
return X, y, y0, nm_landmarks
Import training data and testing data¶
sigma = 5
X_train, y_train, y_train0, nm_landmarks = load2d(test=False,sigma=sigma)
X_test, y_test, _, _ = load2d(test=True,sigma=sigma)
print X_train.shape,y_train.shape, y_train0.shape
print X_test.shape,y_test
Visualize original gray scale image together with heatmap¶
Some of the heatmaps are just black indicating that some landmarks are not recorded (missing), and all the pixcels from this heatmap are zero.
In principle, my deep learning model can accept all-zero heatmap and handle the missing landmarks with no extra effort. Such approach may be approriate if the landmark does not exist in the image: For example, if the left eye is outside of the image, the heatmap for the left eye should be all zero (while the heatmaps of other landmarks from the same image may or may not be all zeros).
However, in this Kaggle's data, I see that all landmarks exist within the image, but for some reasons, some landmark's (x,y) coordinates are not recorded. See plotted images below. So, using all-zero heatmap in training gives misleading infomation to the model; there is no landmark in the image while there is! We should treat these "missing" (x,y)-coordinate landmarks as mis-labeled or contaminated data, and simply do not use such landmarks in training. This can be achieved using weights, and presented later.
Nplot = y_train.shape[3]+1
for i in range(3):
fig = plt.figure(figsize=(20,6))
ax = fig.add_subplot(2,Nplot/2,1)
ax.imshow(X_train[i,:,:,0],cmap="gray")
ax.set_title("input")
for j, lab in enumerate(nm_landmarks[::2]):
ax = fig.add_subplot(2,Nplot/2,j+2)
ax.imshow(y_train[i,:,:,j],cmap="gray")
ax.set_title(str(j) +"\n" + lab[:-2] )
plt.show()
Data augmentation¶
My previous blog showed that it is very important to augment the available training images to improve the model performance on future, validation data.
The code below can do wide range of affine transformation and horizontal flipping. Note that the horizontal flipping in the landmark detection problem needs to make sure that the left-XX label and rand-XX labels are also flipped. In this Kaggle competition, we have 15 landmarks. For example, when horizontal flip happens the left_eye_center (0th landmark) needs to be swapped to the right_eye_center. To keep track on which pairs of landmarks to be swapped, we introduce a dictionary recording the original and new landmark's index:
landmark_order = {"orig" : [0,1,2,3,4,5,6,7,8,9,11,12],
"new" : [1,0,4,5,2,3,8,9,6,7,12,11]}
In my model, I uses weights and horizontal swap will affects the order of weights. This point will become more clear later in the code.
Note: the data augmentation function is not optimized and it sustantially increase the training time :\
Functions¶
from skimage import transform
from skimage.transform import SimilarityTransform, AffineTransform
import random
def transform_img(data,
loc_w_batch=2,
max_rotation=0.01,
max_shift=2,
max_shear=0,
max_scale=0.01,mode="edge"):
'''
data : list of numpy arrays containing a single image
e.g., data = [X, y, w] or data = [X, y]
X.shape = (height, width, NfeatX)
y.shape = (height, width, Nfeaty)
w.shape = (height, width, Nfeatw)
NfeatX, Nfeaty and Nfeatw can be different
affine transformation for a single image
loc_w_batch : the location of the weights in the fourth dimention
[,,,loc_w_batch]
'''
scale = (np.random.uniform(1-max_scale, 1 + max_scale),
np.random.uniform(1-max_scale, 1 + max_scale))
rotation_tmp = np.random.uniform(-1*max_rotation, max_rotation)
translation = (np.random.uniform(-1*max_shift, max_shift),
np.random.uniform(-1*max_shift, max_shift))
shear = np.random.uniform(-1*max_shear, max_shear)
tform = AffineTransform(
scale=scale,#,
## Convert angles from degrees to radians.
rotation=np.deg2rad(rotation_tmp),
translation=translation,
shear=np.deg2rad(shear)
)
for idata, d in enumerate(data):
if idata != loc_w_batch:
## We do NOT need to do affine transformation for weights
## as weights are fixed for each (image,landmark) combination
data[idata] = transform.warp(d, tform,mode=mode)
return data
def transform_imgs(data, lm,
loc_y_batch = 1,
loc_w_batch = 2):
'''
data : list of numpy arrays containing a single image
e.g., data = [X, y, w] or data = [X, y]
X.shape = (height, width, NfeatX)
y.shape = (height, width, Nfeaty)
w.shape = (height, width, Nfeatw)
NfeatX, Nfeaty and Nfeatw can be different
affine transformation for a single image
'''
Nrow = data[0].shape[0]
Ndata = len(data)
data_transform = [[] for i in range(Ndata)]
for irow in range(Nrow):
data_row = []
for idata in range(Ndata):
data_row.append(data[idata][irow])
## affine transformation
data_row_transform = transform_img(data_row,
loc_w_batch)
## horizontal flip
data_row_transform = horizontal_flip(data_row_transform,
lm,
loc_y_batch,
loc_w_batch)
for idata in range(Ndata):
data_transform[idata].append(data_row_transform[idata])
for idata in range(Ndata):
data_transform[idata] = np.array(data_transform[idata])
return(data_transform)
def horizontal_flip(data,lm,loc_y_batch=1,loc_w_batch=2):
'''
flip the image with 50% chance
lm is a dictionary containing "orig" and "new" key
This must indicate the potitions of heatmaps that need to be flipped
landmark_order = {"orig" : [0,1,2,3,4,5,6,7,8,9,11,12],
"new" : [1,0,4,5,2,3,8,9,6,7,12,11]}
data = [X, y, w]
w is optional and if it is in the code, the position needs to be specified
with loc_w_batch
X.shape (height,width,n_channel)
y.shape (height,width,n_landmarks)
w.shape (height,width,n_landmarks)
'''
lo, ln = np.array(lm["orig"]), np.array(lm["new"])
assert len(lo) == len(ln)
if np.random.choice([0,1]) == 1:
return(data)
for i, d in enumerate(data):
d = d[:, ::-1,:]
data[i] = d
data[loc_y_batch] = swap_index_for_horizontal_flip(
data[loc_y_batch], lo, ln)
# when horizontal flip happens to image, we need to heatmap (y) and weights y and w
# do this if loc_w_batch is within data length
if loc_w_batch < len(data):
data[loc_w_batch] = swap_index_for_horizontal_flip(
data[loc_w_batch], lo, ln)
return(data)
def swap_index_for_horizontal_flip(y_batch, lo, ln):
'''
lm = {"orig" : [0,1,2,3,4,5,6,7,8,9,11,12],
"new" : [1,0,4,5,2,3,8,9,6,7,12,11]}
lo, ln = np.array(lm["orig"]), np.array(lm["new"])
'''
y_orig = y_batch[:,:, lo]
y_batch[:,:, lo] = y_batch[:,:, ln]
y_batch[:,:, ln] = y_orig
return(y_batch)
Visualizing augmented images¶
Notice that the image is shifted and also horizontally flipped in random fashion. When horizontal flip happens, the right eye is labeled as left eye and vise versa.
## example image to visualize the data augmentation
iexample = 139
## Show the first 13 heatmaps
Nhm = 10
plt.imshow(X_train[iexample,:,:,0],cmap="gray")
plt.title("original")
plt.axis("off")
plt.show()
Nplot = 5
fig = plt.figure(figsize=[Nhm*2.5,2*Nplot])
landmark_order = {"orig" : [0,1,2,3,4,5,6,7,8,9,11,12],
"new" : [1,0,4,5,2,3,8,9,6,7,12,11]}
count = 1
for _ in range(Nplot):
x_batch, y_batch = transform_imgs([X_train[[iexample]],
y_train[[iexample]]],
landmark_order)
ax = fig.add_subplot(Nplot,Nhm+1,count)
ax.imshow(x_batch[0,:,:,0],cmap="gray")
ax.axis("off")
count += 1
for ifeat in range(Nhm):
ax = fig.add_subplot(Nplot,Nhm + 1,count)
ax.imshow(y_batch[0,:,:,ifeat],cmap="gray")
ax.axis("off")
if count < Nhm + 2:
ax.set_title(nm_landmarks[ifeat*2][:-2])
count += 1
plt.show()
Split data into training and validation data¶
prop_train = 0.9
Ntrain = int(X_train.shape[0]*prop_train)
X_tra, y_tra, X_val,y_val = X_train[:Ntrain],y_train[:Ntrain],X_train[Ntrain:],y_train[Ntrain:]
del X_train, y_train
input_height, input_width = 96, 96
## output shape is the same as input
output_height, output_width = input_height, input_width
n = 32*5
nClasses = 15
nfmp_block1 = 64
nfmp_block2 = 128
IMAGE_ORDERING = "channels_last"
img_input = Input(shape=(input_height,input_width, 1))
# Encoder Block 1
x = Conv2D(nfmp_block1, (3, 3), activation='relu', padding='same', name='block1_conv1', data_format=IMAGE_ORDERING )(img_input)
x = Conv2D(nfmp_block1, (3, 3), activation='relu', padding='same', name='block1_conv2', data_format=IMAGE_ORDERING )(x)
block1 = MaxPooling2D((2, 2), strides=(2, 2), name='block1_pool', data_format=IMAGE_ORDERING )(x)
# Encoder Block 2
x = Conv2D(nfmp_block2, (3, 3), activation='relu', padding='same', name='block2_conv1', data_format=IMAGE_ORDERING )(block1)
x = Conv2D(nfmp_block2, (3, 3), activation='relu', padding='same', name='block2_conv2', data_format=IMAGE_ORDERING )(x)
x = MaxPooling2D((2, 2), strides=(2, 2), name='block2_pool', data_format=IMAGE_ORDERING )(x)
## bottoleneck
o = (Conv2D(n, (input_height/4, input_width/4),
activation='relu' , padding='same', name="bottleneck_1", data_format=IMAGE_ORDERING))(x)
o = (Conv2D(n , ( 1 , 1 ) , activation='relu' , padding='same', name="bottleneck_2", data_format=IMAGE_ORDERING))(o)
## upsamping to bring the feature map size to be the same as the one from block1
## o_block1 = Conv2DTranspose(nfmp_block1, kernel_size=(2,2), strides=(2,2), use_bias=False, name='upsample_1', data_format=IMAGE_ORDERING )(o)
## o = Add()([o_block1,block1])
## output = Conv2DTranspose(nClasses, kernel_size=(2,2), strides=(2,2), use_bias=False, name='upsample_2', data_format=IMAGE_ORDERING )(o)
## Decoder Block
## upsampling to bring the feature map size to be the same as the input image i.e., heatmap size
output = Conv2DTranspose(nClasses, kernel_size=(4,4), strides=(4,4), use_bias=False, name='upsample_2', data_format=IMAGE_ORDERING )(o)
## Reshaping is necessary to use sample_weight_mode="temporal" which assumes 3 dimensional output shape
## See below for the discussion of weights
output = Reshape((output_width*input_height*nClasses,1))(output)
model = Model(img_input, output)
model.summary()
model.compile(loss='mse',optimizer="rmsprop",sample_weight_mode="temporal")
Sample weights¶
We will only include the annotated landmarks to calculate the loss.
To do this, I will define the weight matrix (N_image, height, width, N_landmark). In this weight matrix, if the (x,y)-coordinate of the landmark of the image is recorded, all the height x width pixcels have value 1, and if the landmark is not recorded, all the pixcels have value 0.
Keras acepts the weight matrix to be at most 2 dimensional at this moment (May 2018) with the first dimension corresponding to the image sample. We reshape the weight matrix into the size (N_image, height x width x N_landmark).
def find_weight(y_tra):
'''
:::input:::
y_tra : np.array of shape (N_image, height, width, N_landmark)
:::output:::
weights :
np.array of shape (N_image, height, width, N_landmark)
weights[i_image, :, :, i_landmark] = 1
if the (x,y) coordinate of the landmark for this image is recorded.
else weights[i_image, :, :, i_landmark] = 0
'''
weight = np.zeros_like(y_tra)
count0, count1 = 0, 0
for irow in range(y_tra.shape[0]):
for ifeat in range(y_tra.shape[-1]):
if np.all(y_tra[irow,:,:,ifeat] == 0):
value = 0
count0 += 1
else:
value = 1
count1 += 1
weight[irow,:,:,ifeat] = value
print("N landmarks={:5.0f}, N missing landmarks={:5.0f}, weight.shape={}".format(
count0,count1,weight.shape))
return(weight)
def flatten_except_1dim(weight,ndim=2):
'''
change the dimension from:
(a,b,c,d,..) to (a, b*c*d*..) if ndim = 2
(a,b,c,d,..) to (a, b*c*d*..,1) if ndim = 3
'''
n = weight.shape[0]
if ndim == 2:
shape = (n,-1)
elif ndim == 3:
shape = (n,-1,1)
else:
print("Not implemented!")
weight = weight.reshape(*shape)
return(weight)
Define weights for training, and validation data.
w_tra = find_weight(y_tra)
weight_val = find_weight(y_val)
weight_val = flatten_except_1dim(weight_val)
y_val_fla = flatten_except_1dim(y_val,ndim=3)
## print("weight_tra.shape={}".format(weight_tra.shape))
print("weight_val.shape={}".format(weight_val.shape))
print("y_val_fla.shape={}".format(y_val_fla.shape))
print(model.output.shape)
Training starts here!¶
nb_epochs = 300
batch_size = 32
const = 10
history = {"loss":[],"val_loss":[]}
for iepoch in range(nb_epochs):
start = time.time()
x_batch, y_batch, w_batch = transform_imgs([X_tra,y_tra, w_tra],landmark_order)
# If you want no data augementation, comment out the line above and uncomment the comment below:
# x_batch, y_batch, w_batch = X_tra,y_tra, w_batch
w_batch_fla = flatten_except_1dim(w_batch,ndim=2)
y_batch_fla = flatten_except_1dim(y_batch,ndim=3)
hist = model.fit(x_batch,
y_batch_fla*const,
sample_weight = w_batch_fla,
validation_data=(X_val,y_val_fla*const,weight_val),
batch_size=batch_size,
epochs=1,
verbose=0)
history["loss"].append(hist.history["loss"][0])
history["val_loss"].append(hist.history["val_loss"][0])
end = time.time()
print("Epoch {:03}: loss {:6.4f} val_loss {:6.4f} {:4.1f}sec".format(
iepoch+1,history["loss"][-1],history["val_loss"][-1],end-start))
for label in ["val_loss","loss"]:
plt.plot(history[label],label=label)
plt.legend()
plt.show()
Model performance on example training images¶
The plot shows that when the facial keypoints are not recorded, the estimated model's heatmap is generally low.
y_pred = model.predict(X_tra)
y_pred = y_pred.reshape(-1,output_height,output_width,nClasses)
Nlandmark = y_pred.shape[-1]
for i in range(96,100):
fig = plt.figure(figsize=(3,3))
ax = fig.add_subplot(1,1,1)
ax.imshow(X_tra[i,:,:,0],cmap="gray")
ax.axis("off")
fig = plt.figure(figsize=(20,3))
count = 1
for j, lab in enumerate(nm_landmarks[::2]):
ax = fig.add_subplot(2,Nlandmark,count)
ax.set_title(lab[:10] + "\n" + lab[10:-2])
ax.axis("off")
count += 1
ax.imshow(y_pred[i,:,:,j])
if j == 0:
ax.set_ylabel("prediction")
for j, lab in enumerate(nm_landmarks[::2]):
ax = fig.add_subplot(2,Nlandmark,count)
count += 1
ax.imshow(y_tra[i,:,:,j])
ax.axis("off")
if j == 0:
ax.set_ylabel("true")
plt.show()
Evaluate the performance in (x,y) coordinate¶
Transform heatmap back to the (x,y) coordinate¶
In order to evaluate the model performance in terms of the root mean square error (RMSE) on (x,y) coordiantes, I need to transform heatmap of the landmarks back to the (x,y) coordiantes.
The simplest way would be to use the (x,y) coordiantes of the pixcel with the largest estimated density as the estimated coordinate. In this procedure, however, the estimated (x,y) coordinates are always integers while the true (x,y) coordinates are not necessarily integers. Instead, we may use weighted average of the (x,y) coordinates corresponding to the pixcels with the top "n_points" largest estimated density.
Then question is, how many "n_points" should we use to calculate the weighted average of the coordinates. In the following script, I experiment the effects of the change in "n_points" on the RMSE using training set For the choice of "n_points" I only consider $n^2$ for integer $n$ to allow that selected coordinates to form symetric geometry.
RMSE is calculated in three ways:
- RMSE1: (x,y) coordinates from estimated heatmap VS (x,y) coordinates from true heatmap
- RMSE2: (x,y) coordinates from est heatmap VS true (x,y) coordinates of the landmark
- RMSE3: (x,y) coordinates from true heatmap VS true (x,y) coordinates of the landmark
Ideally, we want to find n_points that returns the smallest RMSE1, RMSE2 and RMSE3.
To reduce the computation time, I will only use the subset of training images for this experiment.
Results¶
The largest "n_points" = $96$x$96$ does not return the smallest RMSE1, RMSE2 or RMSE3. This makes sense because the iamge size is finite and some of the landmarks are at the corner. Taking weighted average across the entire image may results will bias in the coordinate values toward the center of images. This would be the reason why RMSE3 is never 0.
This observation makes me think that it would be better to make the n_points depends on each (image, landmark) combination separately. For example, if the highest density point is at (0,0), then we should not consider large n_points because the estimated density at (-1,-1) would have been large but density in such coordinates are not estimated. On the other hand, if the highest density point is at around the center of the image, then I should consider large n_points. For the simpliciy, I will not implement such procedure, and this would be my future work.
I will use n_points = 25 as it yields the smallest RMSE3.
def get_ave_xy(hmi, n_points = 4, thresh=0):
'''
hmi : heatmap np array of size (height,width)
n_points : x,y coordinates corresponding to the top densities to calculate average (x,y) coordinates
convert heatmap to (x,y) coordinate
x,y coordinates corresponding to the top densities
are used to calculate weighted average of (x,y) coordinates
the weights are used using heatmap
if the heatmap does not contain the probability > Prepare submission¶
Evaluate the model performance on testing images for submission.
y_pred_test = model.predict(X_test) ## estimated heatmap
y_pred_test = y_pred_test.reshape(-1,output_height,output_width,nClasses)
y_pred_test_xy = transfer_target(y_pred_test,thresh=0,n_points=n_points_final) ## estimated xy coord
y_pred_test_xy = pd.DataFrame(y_pred_test_xy,columns=nm_landmarks)
IdLookup = pd.read_csv(os.path.expanduser(FIdLookup))
def prepare_submission(y_pred4,loc):
'''
loc : the path to the submission file
save a .csv file that can be submitted to kaggle
'''
ImageId = IdLookup["ImageId"]
FeatureName = IdLookup["FeatureName"]
RowId = IdLookup["RowId"]
submit = []
for rowId,irow,landmark in zip(RowId,ImageId,FeatureName):
submit.append([rowId,y_pred4[landmark].iloc[irow-1]])
submit = pd.DataFrame(submit,columns=["RowId","Location"])
## adjust the scale
submit["Location"] = submit["Location"]
submit.to_csv(loc,index=False)
print("File is saved at:" + loc)
filename = "result/FCNish_point{:03.0f}.csv".format(n_points_final)
prepare_submission(y_pred_test_xy,filename)